DeepDOF
Intra-operative histopathology
Microscopic evaluation of resected tissue plays a central role in the surgical management of cancer. Because optical microscopes have a limited depth-of-field (DOF), resected tissue is either frozen or preserved with chemical fixatives, sliced into thin sections placed on microscope slides, stained, and imaged to determine whether surgical margins are free of tumor cells—a costly and time- and labor-intensive procedure. Here, we introduce a deep-learning extended DOF (DeepDOF) microscope to quickly image large areas of freshly resected tissue to provide histologic-quality images of surgical margins without physical sectioning.
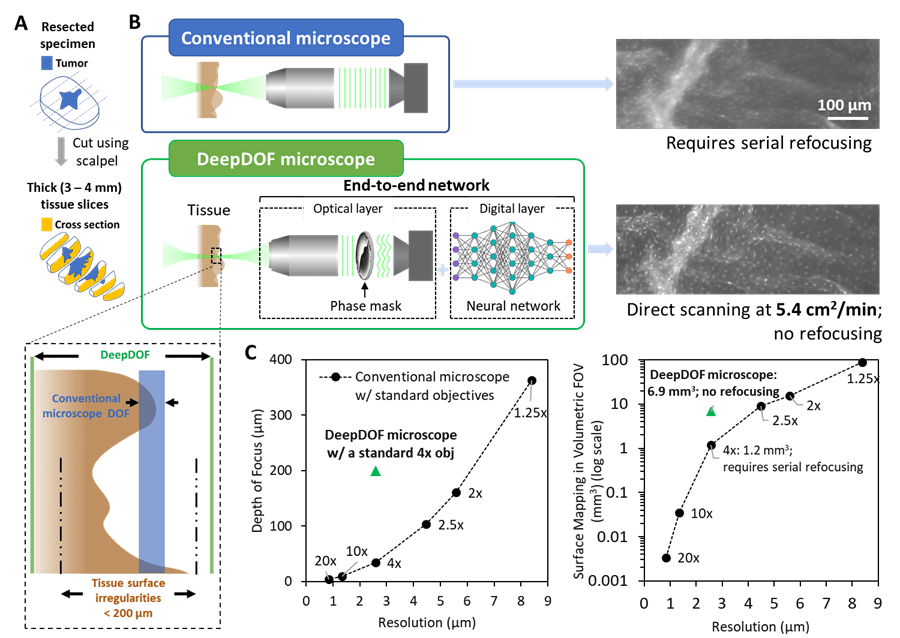
(A) Prior to imaging, the resected specimen is bread-loafed by using a pathology scalpel, and the cross-section surface can be evaluated for tumor-margin assessment. (B) Variations in the surface topology of intact tissue specimens exceed the DOF of a conventional microscope with subcellular resolution. In comparison, with the simple addition of an inexpensive phase mask, the end-to-end optimized DeepDOF microscope allows subcellular imaging of large areas of intact tissue samples at 5.4 cm2/min. (C) Based on a standard 4× objective (obj), the DeepDOF microscope combines wavefront encoding with deep-learning-enabled image reconstruction to significantly improve the DOF and, thus, the volumetric FOV while maintaining subcellular resolution.
End-to-end designed microscope
The DeepDOF microscope consists of a conventional fluorescence microscope with the simple addition of an inexpensive (less than $10) phase mask inserted in the pupil plane to encode the light field and enhance the depth-invariance of the point-spread function. When used with a jointly optimized image-reconstruction algorithm, diffraction-limited optical performance to resolve subcellular features can be maintained while significantly extending the DOF (200 μm). Data from resected oral surgical specimens show that the DeepDOF microscope can consistently visualize nuclear morphology and other important diagnostic features across highly irregular resected tissue surfaces without serial refocusing. With the capability to quickly scan intact samples with subcellular detail, the DeepDOF microscope can improve tissue sampling during intraoperative tumor-margin assessment, while offering an affordable tool to provide histological information from resected tissue specimens in resource-limited settings.
Rapid
More Info
The depth-of-field of the DeepDOF microscope is significantly extended to 200 µm, accommodating for variations in the surface topology of thick cross-sectional tissue slices. Due to its capability to map irregular surfaces in a high-volumetric FOV (6.9 mm³ in DeepDOF microscope vs. 1.2 mm³ in a conventional microscope) with subcellular resolution, the DeepDOF microscope can image large areas of bread-loafed tissue slices without refocusing. We show that using the current economical sample stage, DeepDOF can scan large specimens at 5.4 cm²/min. The robust image reconstruction only takes an average of 0.5 s/mm² using the TensorFlow package, and its speed can be further optimized.
Low-cost
More Info
The DeepDOF microscope is built on a conventional epifluorescence microscope with the simple addition of an inexpensive (less than $10) phase mask, and its low cost (less than $6,000) allows potential adoption in communities in low-resource settings.
Clinically significant features
More Info
By using the DeepDOF microscope, diagnostic features similar to those in conventional H&E slides with subcellular detail can be directly visualized from an intact tissue sample, circumventing tissue alterations resulting from processing
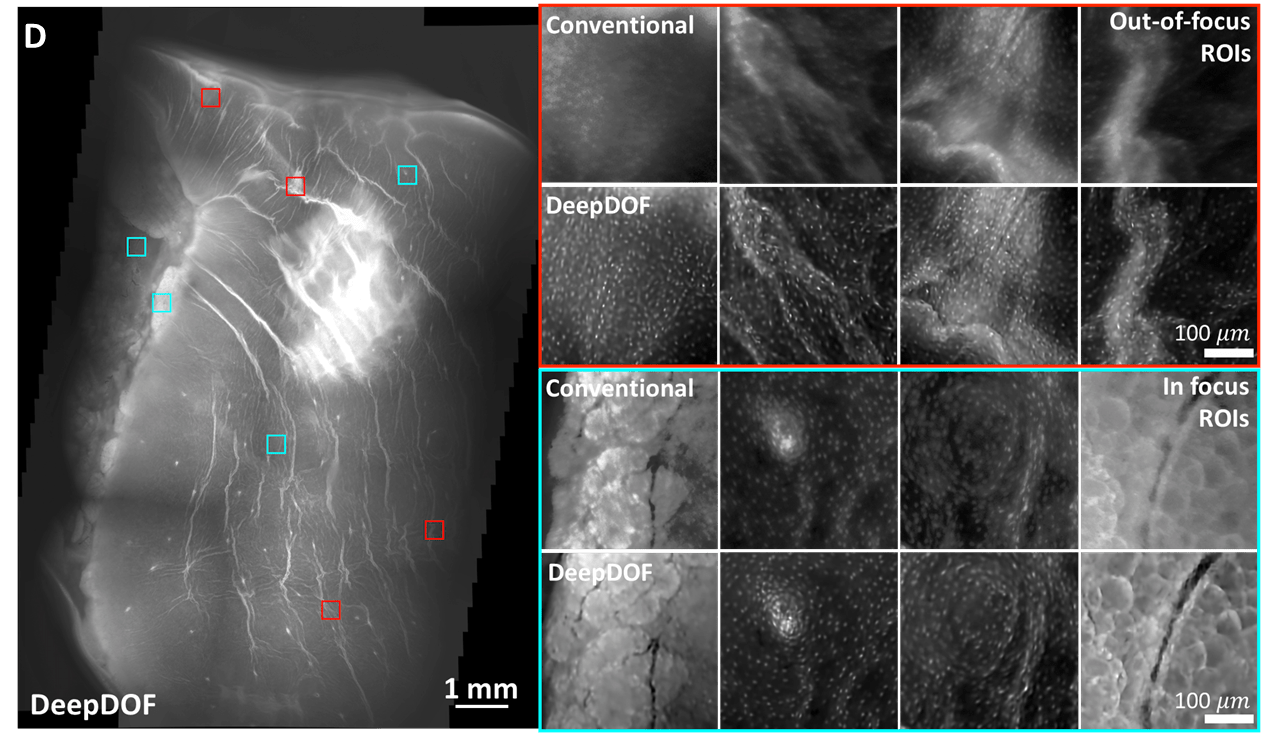
The DeepDOF image clearly visualizes subcellular features, primarily individual nuclei that are evenly distributed on the esophageal wall. Multiple ROIs are shown in regions where the conventional microscope images are out of focus (red) and in focus (cyan).
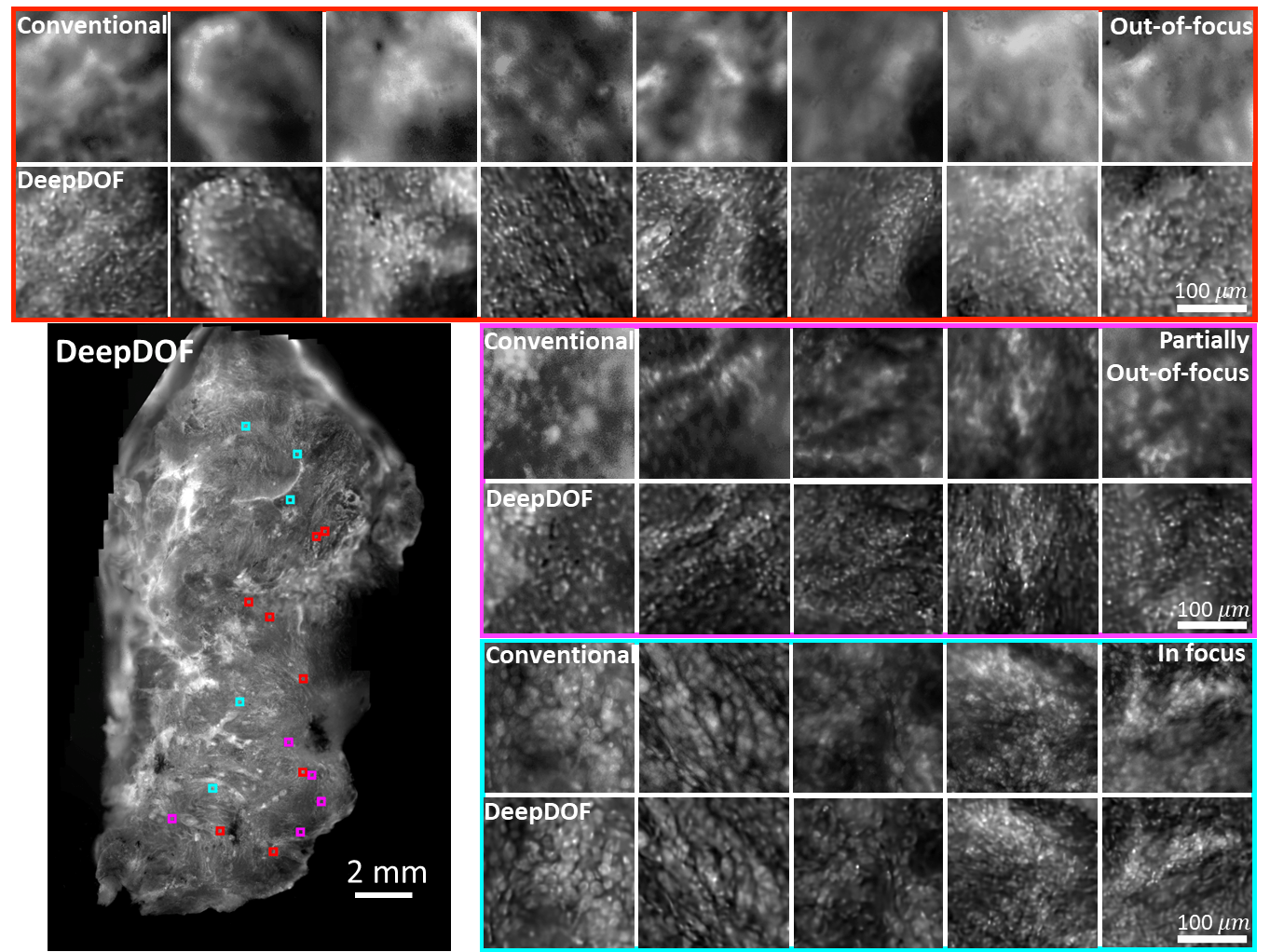
Images acquired with the DeepDOF microscope and a conventional microscope (Olympus 4×, 0.13 NA) from a large surgically resected specimen from the oral cavity. Three groups of ROIs show regions where the conventional microscope images are significantly out of focus (red), partially out of focus (magenta), and in focus (cyan). The contrast of the conventional images is matched to show the effects of defocus blur.
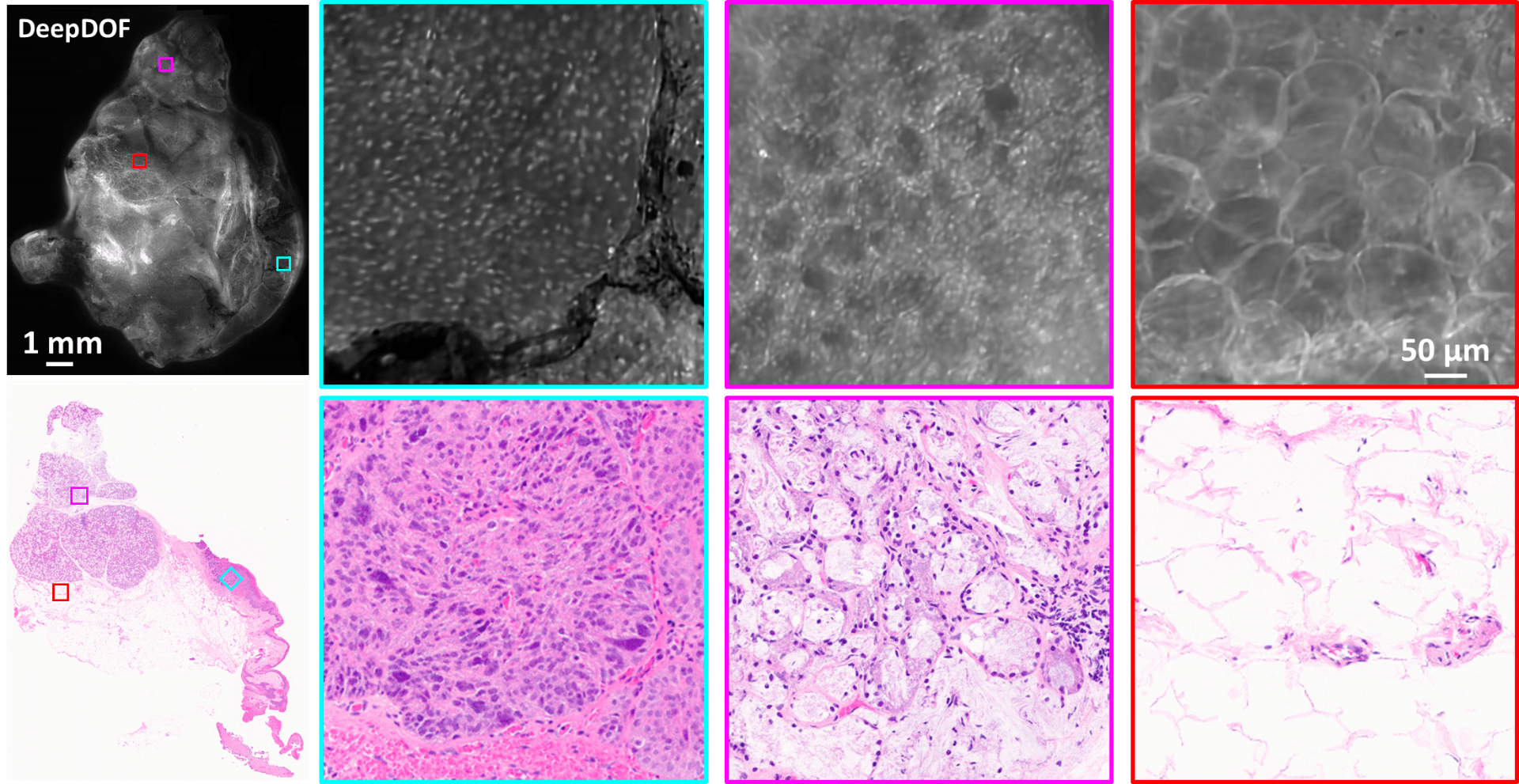
The DeepDOF microscope visualizes important diagnostic features in large proflavine-stained surgical specimen with subcellular resolution. Three selected ROIs across the DeepDOF image of the specimen reveal a nest of neoplastic cells in the squamous epithelium (cyan), glandular patterns in salivary tissue (magenta), and adipose cells (red), respectively. Image features were confirmed by the gold standard of histopathology.
People

Mary Jin

Yubo Tang

Yicheng Wu

Jackson B. Coole

Melody T. Tan

Xuan Zhao
Hawraa Badaoui

Jacob T. Robinson

Michelle D. Williams

Ann M. Gillenwater

Rebecca R. Richards-Kortum

Ashok Veeraraghavan
